Cluster Analysis by Multidimensional Scaling using Systat 9
Input data are soil analyses from Daisy property, Montana (-80 mesh fraction, aqua-regia digestion, 0.5 gram aliquot). Appropriate data transformations (to create Normal distributions) have already been selected and applied.
|
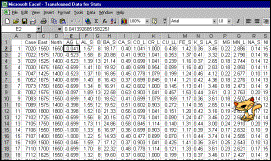
Input data set |
Step
1: Standardize the data (subtract mean and divide by standard deviation) |
|
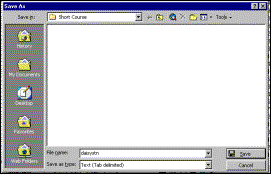
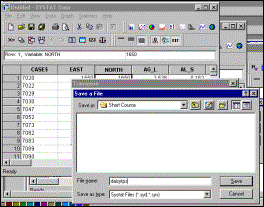
Step
2: Save as a tab-delimited file for Systat |
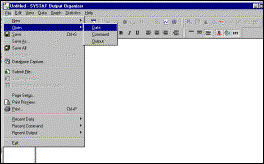
Step
3: Run Systat |
|
Step
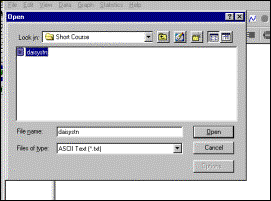
4: Open the data file |
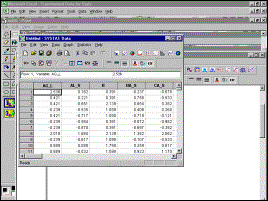
Systat data window |
|
Step
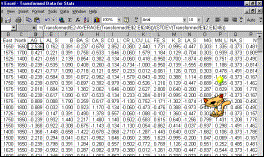
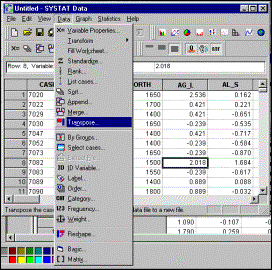
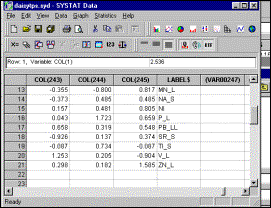
5: Transpose input data (convert rows to columns, and vice versa) |
Step
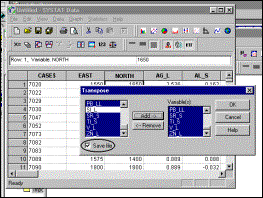
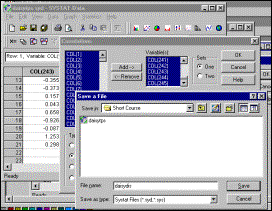
6: Select elements to be included in transposed matrix, and specify that
output file must be saved |
|
Step
7: Specify name of output file |
Lower right-hand corner
of output file in Systat data window |
|
Step
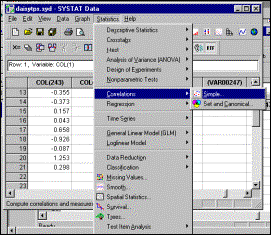
8: Calculate inter-column (sample) distances using Statistics à Correlation
option |
Step
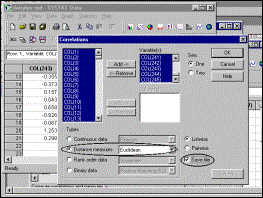
9: Specify method of distance measurement and that output data should be
saved |
|
Step
10: Specify name of output file |
Lower right hand corner
of triangular distance matrix (may take several minutes to generate) |
|
Step
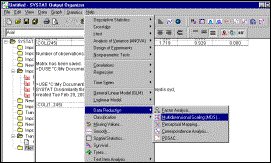
11: Load file of distances (created in previous step) and select
Statistics à Data
Reduction à
Multidimensional Scaling option |
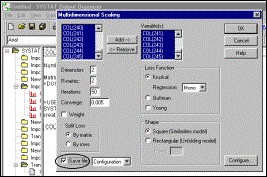
Step
12: Specify optimisation method (Systat defaults are acceptable), and that
output should be saved |
|
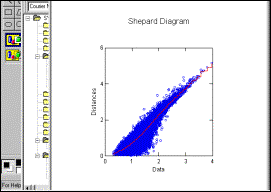
Shepard diagram shows
relationship between intersample distances in multielement (x-axis) and
2-dimensional (y-axis) space |
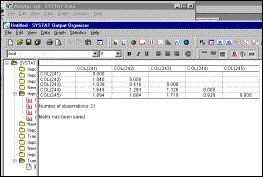
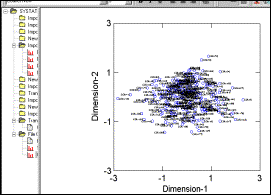
MDS configurations are
output to screen as well as to file |
|
Plot created by Systat.
It is not very suitable for cluster identification |
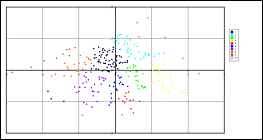
Plot created in MS-Excel
and MS-Paint from output file of MDS configurations |